

The goal of grateful is to make it very easy to cite R and the R packages used in any analyses, so that package authors receive their deserved credit. By calling a single function, grateful will scan the project for R packages used and generate a BibTeX file containing all citations for those packages.
grateful can then generate a new document with citations in the desired output format (Word, PDF, LaTeX, HTML, Markdown). These references can be formatted for a specific journal, so that we can just paste them directly into our manuscript or report.
Alternatively, we can use grateful directly within an Rmarkdown or Quarto document. In this case, a paragraph containing in-text citations of all used R packages will (optionally) be inserted into the Rmarkdown/Quarto document, and these packages will be included in the reference list when rendering.
You can install the stable release of {grateful} from CRAN:
install.packages("grateful")Or the latest development version from R-universe:
install.packages("grateful", repos = c("https://pakillo.r-universe.dev", "https://cloud.r-project.org"))Or from GitHub:
# install.packages("remotes")
remotes::install_github("Pakillo/grateful")grateful can be used in one of two ways:
to generate a ‘citation report’ listing each package and their citations
to build citation keys to incorporate into an existing R Markdown or Quarto document.
Imagine a project where we are using the packages: dplyr, ggplot2, vegan and lme4. We want to collect all the citations listed for these packages, as well as a citation for base R (and for RStudio, if applicable).
Calling cite_packages() will scan the project, find
these packages, and generate a document with formatted citations.
library(grateful)cite_packages(out.dir = ".") # save report to working directory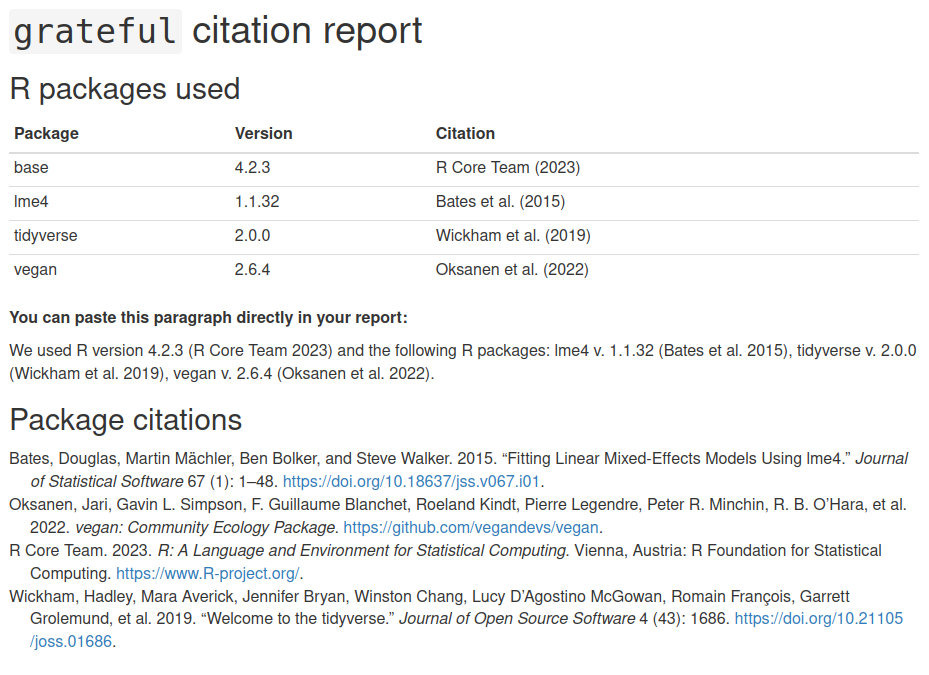
This document can also be a Word or LaTeX document, PDF file,
markdown file, or left as the source Rmarkdown file using
out.format:
cite_packages(out.format = "docx", out.dir = ".")We can specify the citation style for a particular journal using
citation.style.
cite_packages(citation.style = "peerj", out.dir = ".")In all cases a BibTeX (.bib) file with all package citations will be saved to disk.
If you are building a document in RMarkdown or Quarto and want to cite R packages, grateful can automatically generate a BibTeX file and ensure these packages are cited in the appropriate format (see template Rmarkdown and Quarto documents).
First, include a reference to the BibTeX file in your YAML header.
bibliography: grateful-refs.bib(Note: You can reference multiple BibTeX files, if needed)
bibliography:
- document_citations.bib
- grateful-refs.bibThen call cite_packages(output = "paragraph") within a
code chunk (block or inline) to automatically include a paragraph
mentioning all the used packages, and include their references in the
bibliography list.
```{r}
cite_packages(output = "paragraph", out.dir = ".")
```We used R version 4.2.3 [@base] and the following R packages: lme4 v. 1.1.32 [@lme4], tidyverse v. 2.0.0 [@tidyverse], vegan v. 2.6.4 [@vegan].
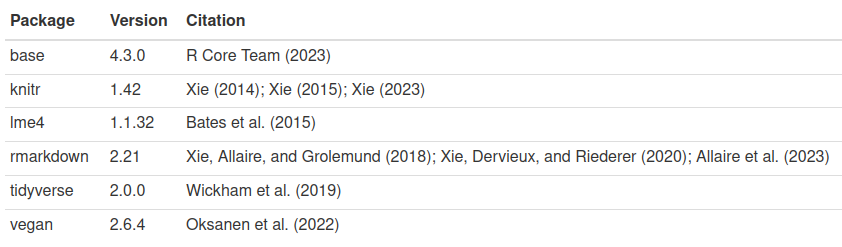
Alternatively, you can get a table with package name, version, and
citations, using output = 'table':
```{r }
pkgs <- cite_packages(output = "table", out.dir = ".")
knitr::kable(pkgs)
```
If you want the references to appear in a particular format, you can specify the citation style in the YAML header:
bibliography: grateful-refs.bib
csl: peerj.cslAlternatively, you can cite particular packages using the citation
keys generated by grateful, as with any other BibTeX
reference, or just include citations in the References section, using
the function nocite_references(). See the package help and
the RMarkdown
cookbook for more details.
Use scan_packages
scan_packages()
pkg version
1 badger 0.2.5
2 base 4.5.1
3 desc 1.4.3
4 knitr 1.50
5 mgcv 1.9.3
6 pkgdown 2.1.3
7 remotes 2.5.0
8 renv 1.1.5
9 rmarkdown 2.29
10 testthat 3.2.3
11 tidyverse 2.0.0
12 visreg 2.7.0If you just want to get all package references in a BibTeX file, you
can call get_pkgs_info(). Besides printing a table with
package info, it will also save a BibTeX file with references. By
default, the file will be called grateful-refs.bib, but you
can change that (see function help).
If you want to get the BibTeX references for a few specific packages:
get_pkgs_info(pkgs = c("remotes", "renv"), out.dir = getwd())
#> pkg version citekeys
#> 1 remotes 2.5.0 remotes
#> 2 renv 1.1.5 renvIf you use one or several packages from the tidyverse, you can choose to cite the ‘tidyverse’ rather than the individual packages:
cite_packages(cite.tidyverse = TRUE)Most R packages also depend on other packages. To include those
package dependencies in your citations, rather than just the packages
you called directly, use dependencies = TRUE:
cite_packages(dependencies = TRUE)Some R packages wrap core external software that should perhaps be
cited too. For example, rjags
is an R wrapper to the JAGS software written in
C++. Ideally, R packages wrapping core external software will include
them in their CITATION file. But otherwise, we can investigate external
software requirements of our used packages, e.g. using
remotes:
remotes::system_requirements(package = c("rjags"), os = "ubuntu-20.04")
#> [1] "apt-get install -y jags"Citing software is pretty much like citing papers. Authors have to decide what to cite in each case, which depends on research context.
As written in the Software Citation Principles paper (Smith et al. 2016):
The software citation principles do not define what software should be cited, but rather how software should be cited. What software should be cited is the decision of the author(s) of the research work in the context of community norms and practices, and in most research communities, these are currently in flux. In general, we believe that software should be cited on the same basis as any other research product such as a paper or book; that is, authors should cite the appropriate set of software products just as they cite the appropriate set of papers, perhaps following the FORCE11 Data Citation Working Group principles, which state, “In scholarly literature, whenever and wherever a claim relies upon data, the corresponding data should be cited”
And these are the guidelines from the Software Citation Checklist:
You should cite software that has a significant impact on the research outcome presented in your work, or on the way the research has been conducted. If the research you are presenting is not repeatable without a piece of software, then you should cite the software. Note that the license or copyright of the software has no bearing on whether you should cite it.
This might include:
Software (including scripts) you have written yourself to conduct the research presented. A software framework / platform upon which the software you wrote to conduct the research relies. Software packages, plugins, modules and libraries you used to conduct your research and that perform a critical role in your results. Software you have used to simulate or model phenomena/systems. Specialist software (which is not considered commonplace in your field) used to prepare, manage, analyse or visualise data. Software being evaluated or compared as part of the research presented Software that has produced analytic results or other output, especially if used through an interface.
In general, you do not need to cite:
Software packages or libraries that are not fundamental to your work and that are a normal part of the computational and scientific environment used. These dependencies do not need to be cited outright but should be documented as part of the computational workflow for complete reproducibility. Software that was used during the course of the research but had no impact on research results, e.g. word processing software, backup software.
Apart from citing the software most relevant to the particular
research/analysis performed, I think it is good idea to record the
entire computational environment elsewhere, e.g. using
sessionInfo() or
sessioninfo::session_info().
Some packages include more than one citation (e.g. knitr,
mgcv).
grateful will include all those citations by default, as it
is impossible to decide automatically which citations should be included
in each case. The user may manually remove citations from the produced
reference list after calling cite_packages. If using Quarto
or Rmarkdown, we can generate the citation paragraph and manually remove
the unwanted references so they will not appear cited.
For example, mgcv package provides multiple references
to be cited:
citation("mgcv")To choose just one of them to be cited, we could generate a citation
paragraph using cite_packages
cite_packages("paragraph", out.dir = ".")
And then manually remove the unwanted citation keys, leaving just those we want to cite:

When rendering the Rmarkdown or Quarto document, only the chosen references will be cited.
Before running grateful you might want to run lintr::unused_import_linter,
funchir::stale_package_check
or annotater
to check for unused packages before citing them.
If getting an error like
Error in (function (pkg, lib.loc = NULL): there is no package called...,
that means that some of your scripts is loading a package that is no
longer available in your computer, so {grateful} cannot grab its
citation. To fix this, there are several options. First, you could omit
that package (or those packages, if more than one) from {grateful}
citations using
cite_packages(omit = c("package1", "package2"). Second, you
could set a .renvignore file to ignore particular files or
folders (see instructions here).
Third, try checking
if that package is still needed for your project and you want to
cite it; otherwise remove or comment that line where the package is
loaded. If you still use and want to cite that package, install it, and
then run cite_packages again. Finally, you could use the
argument skip.missing = TRUE to skip those missing packages
from the citation list.
When a project includes many used packages (or files),
renv may issue a warning. Use
options(renv.config.dependencies.limit = 10000) to overcome
the warning and scan the project for all packages used. Alternatively,
use .renvignore to ignore certain files or folders (see
renv help).
Use a .renvignore file to ignore certain files or
folders (see renv help).
Here
are example Quarto
and Rmarkdown
documents showing how to generate a separate bibliography for R packages
(different from the main bibliography). This requires installing the multibib
extension first.
To cite the dependencies of an R package as stated in its DESCRIPTION
file, use
pkgs = c("Depends", "Imports", "Suggests", "LinkingTo") or
a combination of them to obtain the desired type of dependencies.
For example, these are {grateful} package ‘Imports’ and ‘Suggests’:
cite_packages(output = "table", out.dir = ".", pkgs = c("Imports", "Suggests"))
#> Package Version Citation
#> 1 curl <NA> @curl
#> 2 desc <NA> @desc
#> 3 knitr <NA> @knitr2014; @knitr2015; @knitr2025
#> 4 remotes <NA> @remotes
#> 5 renv <NA> @renv
#> 6 rmarkdown <NA> @rmarkdown2018; @rmarkdown2020; @rmarkdown2024
#> 7 rstudioapi <NA> @rstudioapi
#> 8 testthat >= 3.0.0 @testthat
#> 9 utils <NA> @utilsTo also include the dependencies from those packages, use
dependencies = TRUE:
out <- cite_packages(output = "table", out.dir = ".", pkgs = c("Imports", "Suggests"),
dependencies = TRUE)
head(out)
#> Package Version Citation
#> 1 base64enc <NA> @base64enc
#> 2 brio <NA> @brio
#> 3 bslib <NA> @bslib
#> 4 cachem <NA> @cachem
#> 5 callr <NA> @callr
#> 6 cli <NA> @clicite_packages includes a few arguments
(text.start, text.pkgs and
text.RStudio) to allow the user to customise the language
of the citation paragraph.
For example, to produce a citation paragraph in Spanish:
cite_packages(output = "paragraph", out.dir = ".",
text.start = "Para desarrollar este trabajo se utilizó",
text.pkgs = "y los siguientes paquetes")"Para desarrollar este trabajo se utilizó R version 4.5.1 [@base] y los siguientes paquetes: badger v. 0.2.5 [@badger], desc v. 1.4.3 [@desc], knitr v. 1.50 [@knitr2014; @knitr2015; @knitr2025], mgcv v. 1.9.3 [@mgcv2003; @mgcv2004; @mgcv2011; @mgcv2016; @mgcv2017], pkgdown v. 2.1.3 [@pkgdown], remotes v. 2.5.0 [@remotes], renv v. 1.1.5 [@renv], rmarkdown v. 2.29 [@rmarkdown2018; @rmarkdown2020; @rmarkdown2024], testthat v. 3.2.3 [@testthat], tidyverse v. 2.0.0 [@tidyverse], visreg v. 2.7.0 [@visreg]."
Or in German:
cite_packages(output = "paragraph", out.dir = ".",
text.start = "Wir verwendeten die",
text.pkgs = "und die folgenden R-Pakete")"Wir verwendeten die R version 4.5.1 [@base] und die folgenden R-Pakete: badger v. 0.2.5 [@badger], desc v. 1.4.3 [@desc], knitr v. 1.50 [@knitr2014; @knitr2015; @knitr2025], mgcv v. 1.9.3 [@mgcv2003; @mgcv2004; @mgcv2011; @mgcv2016; @mgcv2017], pkgdown v. 2.1.3 [@pkgdown], remotes v. 2.5.0 [@remotes], renv v. 1.1.5 [@renv], rmarkdown v. 2.29 [@rmarkdown2018; @rmarkdown2020; @rmarkdown2024], testthat v. 3.2.3 [@testthat], tidyverse v. 2.0.0 [@tidyverse], visreg v. 2.7.0 [@visreg]."
citation("grateful")
To cite package 'grateful' in publications use:
Rodriguez-Sanchez F, Jackson C (2025). _grateful: Facilitate citation
of R packages_. <https://pakillo.github.io/grateful/>.
A BibTeX entry for LaTeX users is
@Manual{,
title = {grateful: Facilitate citation of {R} packages},
author = {Francisco Rodriguez-Sanchez and Connor P. Jackson},
year = {2025},
url = {https://pakillo.github.io/grateful/},
}Citation keys are not guaranteed to be preserved when regenerated, particularly when packages are updated. This instability is not an issue when citations are used programmatically, as in the example above. But if references are put into the text manually, they may need to be updated periodically.