

The package’s website is available here
In versions previous 0.4.4, we exported a class using c++ to do
spatial indexing. It caused bugs on CRAN server and was removed because
it was not significantly faster than sf::st_join.
We reworked (version 0.4.4) the functions to calculate K functions on
network:
kfunctions,kfunctions.mc,cross_kfunctions,cross_kfunctions.mc,k_nt_functions
and k_nt_functions.mc. We are still working on the
spatio-temporal version of the cross K functions. Also, the current
implementation of the G function is quite literal and we will try to
replace it by a more interesting one using kernel density for
estimation.
The version 0.4.4 has modifications in the arguments of the functions
bw_cvl_calc, bw_cvl_calc.mc,
bw_cv_likelihood_calc,
bw_cv_likelihood_calc,bw_tnkde_cv_likelihood_calc.mc.
The parameters about bandwidths range and step were replaced by a unique
parameter requiring and ordered vector of bandwidths.
Code from previous version need to be modified accordingly.
A very interesting package has been published with
cppRouting. It allows for extremely fast shortest path
calculation on network. We are slowly integrating it in
spNetwork. The changes are not visible for the users
Considering that rgeos and maptools will be
deprecated soon, we are moving to sf! This requires some adjustment in
the code and the documentation. The development version and the releases
on CRAN are now using sf. Please, report any bug or error
in the documentation.
To install the previous version using sp,
rgeos and maptools, you can run the following
command:
devtools::install_github("JeremyGelb/spNetwork", ref = "a3bc982")Note that all the new developments will use sf and you
should switch as soon as possible.
Because of a new CRAN policy, it is not possible anymore to read data
in gpkg if they are stored in the user library. On Debian systems, this
library is now mounted as read-only for checking. All the datasets
provided by spNetwork are now stored as .rda file, and can be loaded
with the function data.
This package can be used to perform several types of analysis on geographical networks. This type of network have spatial coordinates associated with their nodes. They can be directed or undirected. In the actual development version the implemented methods are:
network_knn).Calculation on network can be long, efforts were made to reduce computation time by implementing several functions with Rcpp and RcppArmadillo and by using multiprocessing when possible.
you can install the CRAN version of this package with the following code in R.
install.packages("spNetwork")To use all the new features before they are available in the CRAN version, you can download the development version.
devtools::install_github("JeremyGelb/spNetwork")The packages uses mainly the following packages in its internal structure :
We provide here some short examples of several features. Please, check the vignettes for more details.
library(spNetwork)
library(tmap)
library(sf)
# loading the dataset
data(mtl_network)
data(bike_accidents)
# generating sampling points at the middle of lixels
samples <- lines_points_along(mtl_network, 50)
# calculating densities
densities <- nkde(lines = mtl_network,
events = bike_accidents,
w = rep(1,nrow(bike_accidents)),
samples = samples,
kernel_name = "quartic",
bw = 300, div= "bw",
method = "discontinuous",
digits = 2, tol = 0.1,
grid_shape = c(1,1),
max_depth = 8,
agg = 5, sparse = TRUE,
verbose = FALSE)
densities <- densities*1000
samples$density <- densities
tm_shape(samples) +
tm_dots(col = "density", size = 0.05, palette = "viridis",
n = 7, style = "kmeans")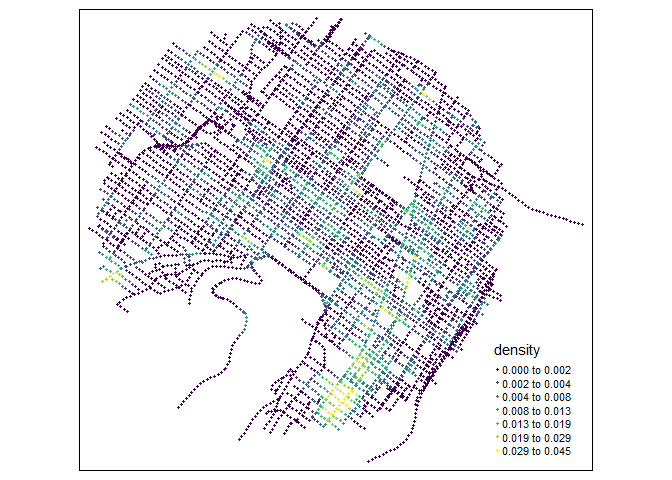
An extension for spatio-temporal dataset is also available Temporal Network Kernel Density Estimate
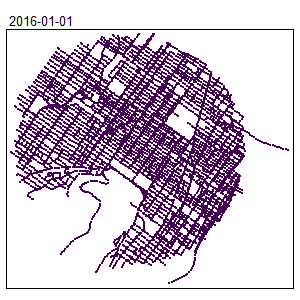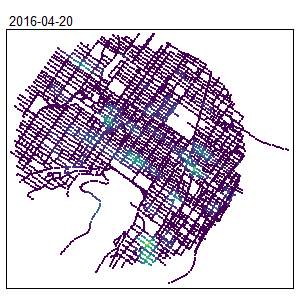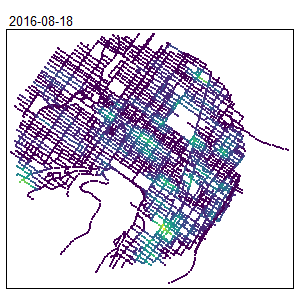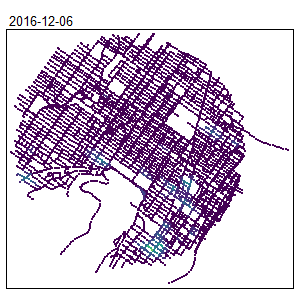
library(spdep)
# creating a spatial weight matrix for the accidents
listw <- network_listw(bike_accidents,
mtl_network,
mindist = 10,
maxdistance = 400,
dist_func = "squared inverse",
line_weight = 'length',
matrice_type = 'W',
grid_shape = c(1,1),
verbose=FALSE)
# using the matrix to find isolated accidents (more than 500m)
no_link <- sapply(listw$neighbours, function(n){
if(sum(n) == 0){
return(TRUE)
}else{
return(FALSE)
}
})
bike_accidents$isolated <- as.factor(ifelse(no_link,
"isolated","not isolated"))
tm_shape(mtl_network) +
tm_lines(col = "black") +
tm_shape(bike_accidents) +
tm_dots(col = "isolated", size = 0.1,
palette = c("isolated" = "red","not isolated" = "blue"))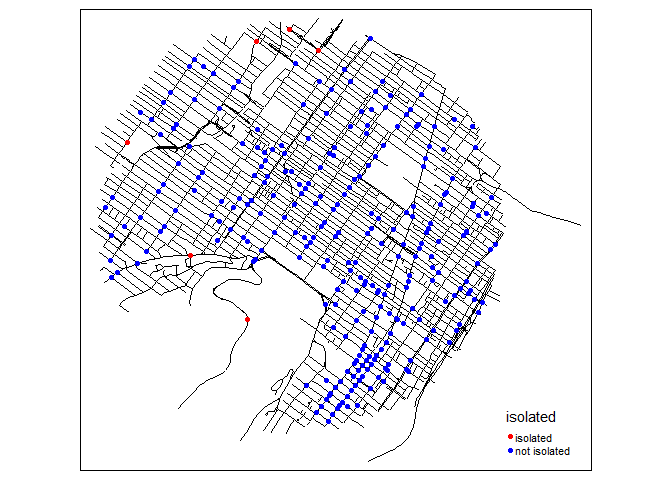
Note that you can use this in every spatial analysis you would like to perform. With the converter function of spdep (like listw2mat), you can convert the listw object into regular matrix if needed
# loading the data
data(main_network_mtl)
data(mtl_theatres)
# calculating the k function
kfun_theatre <- kfunctions(main_network_mtl, mtl_theatres,
start = 0, end = 5000, step = 50,
width = 1000, nsim = 50, resolution = 50,
verbose = FALSE, conf_int = 0.05)
kfun_theatre$plotg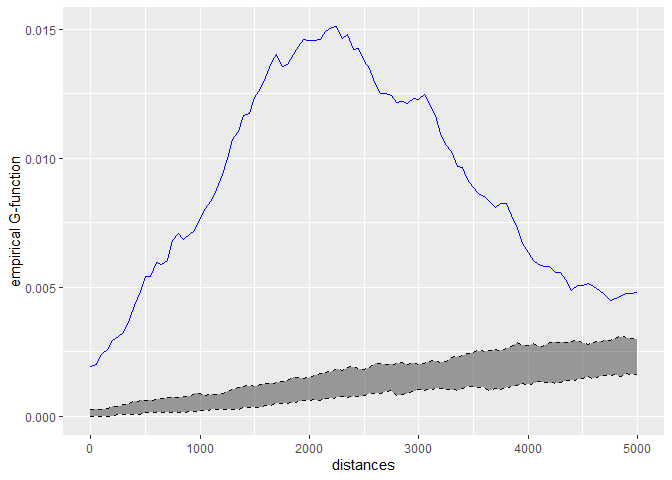
New methods will be probably added in the future, but we will focus on performance for the next release. Do no hesitate to open an issue here if you have suggestion or if you encounter a bug.
Features that will be added to the package in the future:
sf objects rather than sp
(rgeos and maptools will be deprecated in
2023). This work is undergoing, please report any bug or error in the
new documentation.If you encounter a bug when using spNetwork, please open an issue here. To ensure that the problem is quickly identified, the issue should follow the following guidelines:
To contribute to spNetwork, please follow these guidelines.
Please note that the spNetwork project is released with
a Contributor
Code of Conduct. By contributing to this project, you agree to abide
by its terms.
An article presenting spNetwork and NKDE has been
accepted in the RJournal!
Gelb Jérémy (2021). spNetwork, a package for network kernel density estimation. The R Journal. https://journal.r-project.org/archive/2021/RJ-2021-102/index.html.
You can also cite the package for other methods:
Gelb Jérémy (2021). spNetwork: Spatial Analysis on Network. https://jeremygelb.github.io/spNetwork/.
spNetwork is licensed under GPL2
License.